- Index
- Manual Downloads(PDF)
Japanese
Japanese (updated 9/2012)
Japanese (updated 9/2015)
- Introduction to Glycobiology
e-learning ツール
|
DrawRINGS
-
What is DrawRINGS?
DrawRINGS is a web-based tool that searches for already-known glycan structures that are similar to the glycan structure being drawn by using the mouse.
It can output KCF from the glycan structure and can compare the structures by using a specific Glycan Score Matrix.
It can also take a glycan structure in KCF format as input, through the file browser field below DrawRINGS.
-
Usage
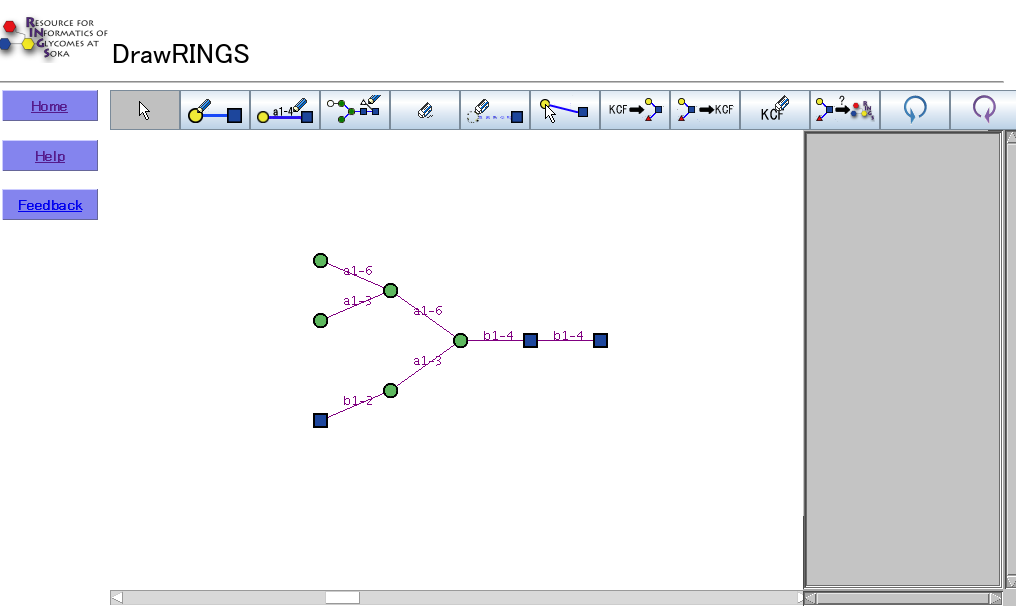
There are nine buttons, a drawing area, and a text area in the drawing interface.
Each button provides a particular functionality, described below..
-
Draw Node
The selection menu of the monosaccharide names appears when the drawing area is clicked, and the selected monosaccharide will be drawn.
Moreover, a pop-up window to input a monosaccharide name appears when 'Other' is selected such that names of entities not appearing in the list can be entered.
-
Draw Edge
Two monosaccharides can be linked by clicking on the corresponding monosaccharides.
The selection menu of link information appears when an edge is clicked.
Moreover, the pop-up menu to input the edge name appears when pushing 'Other' such that link information other than those in the list can be drawn.
-
Core structure
Core structures include the common N-glycan core structures, including bisecting GlcNAcs and core fucosylation, O-glycan structures, GAGs, and common motifs such as Lewis structures, etc.
-
Clear ALL
All glycans and edges are deleted.
-
Erase Node
By clicking one monosaccharide, the selected monosaccharide and corresponding edges will be deleted.
-
Move Node
A selected monosaccharide can be moved by dragging it.
-
Draw KCF
The structure corresponding to the KCF in the text area will be drawn on the canvas.
-
KCF Text out
The structure being drawn is output into KCF.
-
Text Clear
The KCF in the text area is deleted.
-
Run query
The pop-up menu to select the score matrix will appear, and the RINGS database of glycan structures will be searched using the KCaM exact-match algorithm to retrieve the most similar structures. If a score matrix is selected, the algorithm will use the values from the score matrix to weight the linkages appropriately.
-
Undo
The last action performed on the canvas is undone.
-
Redo
The last action undone is redone.
-
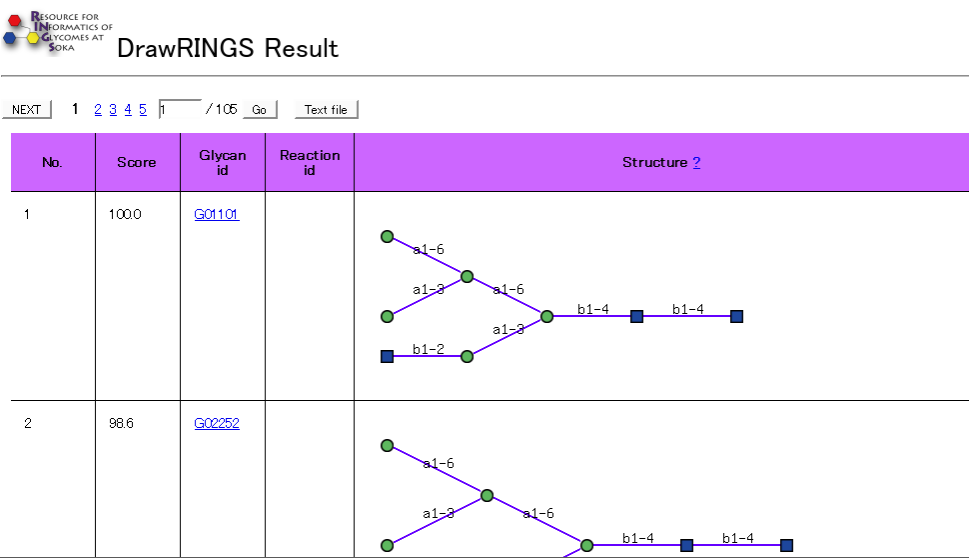
Result
When "Run query" is executed, the result othe structural comparison is displayed on a new page.
Corresponding glycan IDs to the input structure, reaction IDs that relate to the glycan and the images of the glycan structure is displayed in the result.
Information on each sugar chain can be obtained by clicking each link.
(Top)
Legend
These are the symbols of the monosaccharides used in RINGS. This follows the standard set forth by the Consortium for Functional Glycomics (CFG).
|
Symbol
|
Abbrev.
|
Sugar name
|
Symbol
|
Abbrev.
|
Sugar name
|

|
Fuc
|
Fucose
|

|
Gal
|
Galactose
|

|
GalA
|
Galacturonic acid
|

|
GalN
|
Galactosamine
|

|
GalNAc
|
N-acetylgalactosamine
|

|
Glc
|
Glucose
|

|
GlcA
|
Glucuronic acid
|

|
GlcN
|
Glucosamine
|

|
GlcNAc
|
N-acetylglucosamine
|

|
Ido
|
Idose
|

|
Kdn
|
2-keto-3-deoxy-nonulosonic acid
|

|
Man
|
Mannose
|

|
ManA
|
Mannuronic acid
|

|
ManN
|
Mannosamine
|

|
ManNAc
|
N-acetylmannosamine
|

|
NeuAc
|
N-acetylneuraminic acid
|

|
NeuGc
|
N-glycolyl-neuraminic acid
|

|
Xyl
|
Xylose
|
(Top)
GlycanMiner
-
Glycan Miner Overview
Glycan Miner is a tool for mining alpha-closed frequent subtrees from a set of glycan structures, based on the work by Hashimoto et al., 2008. alpha-closed frequent subtrees are defined as those substructures within the input that frequently appear within a specified range. The minimum frequency of appearance must be specified by the user via the minsup parameter. Those substructures that appear in at least minsup number of input glycan structures will be included in the output. Furthermore, in order to avoid too many similar substructures in the results, the alpha parameter is used to specify the amount of overlap, and it is defined by the following equation: support(T') < max(alpha x support(T), minsup), where support(T) refers to the number of glycan structures in which substructure T appears, and substructure T is a subtree of substructure T'.
-
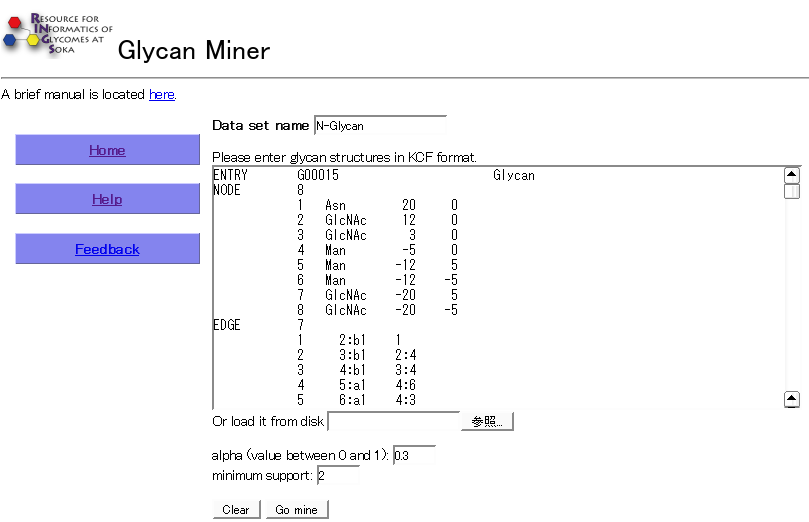
Usage
There is one major text field for entering the glycan structures in KCF format. This input can also be provided in a text file which can be loaded directly. A value between 0 and 1 should be specified for the alpha parameter, and the minimum number by which the resulting substructure should appear should be specified for minimum support.
-
Results
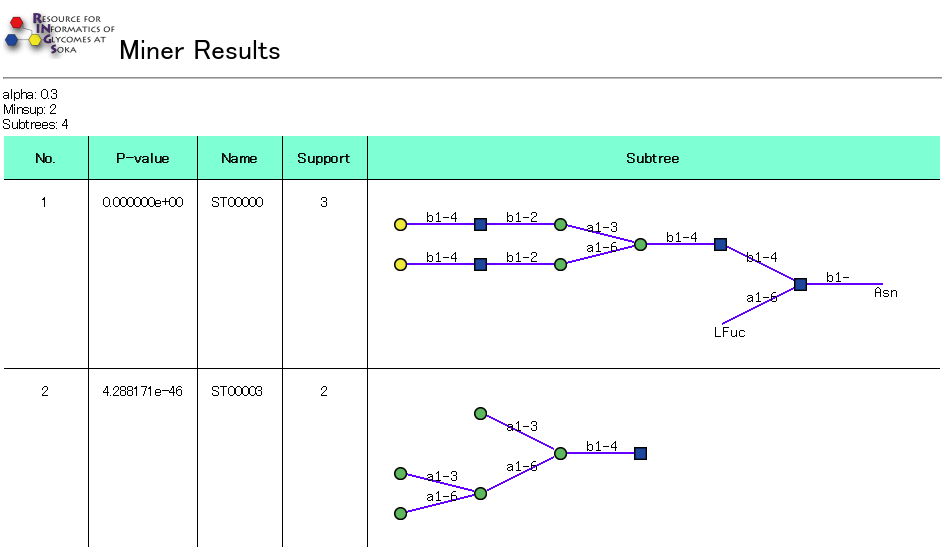
Note that it may take a few minutes for the results to be displayed as much computation is required. We are currently working on optimizing this process.
In the results, each alpha-closed frequent subtree is ranked according to p-value, which is computed by comparing the support of a similar structure with the same components but randomized. The actual value of the structure's support is also displayed, along with the figure of the structure itself. A name to label each structure is also given.
(Top)
GPP
-
Glycan Pathway Predictor (GPP) Overview
Glycan Pathway Predictor (GPP) is a tool for dynamically computing the N-glycan biosynthesis pathway from a given glycan structure, based on the paper by Krambeck et al., 2005, which was later improved in Krambeck et al., 2009.
-
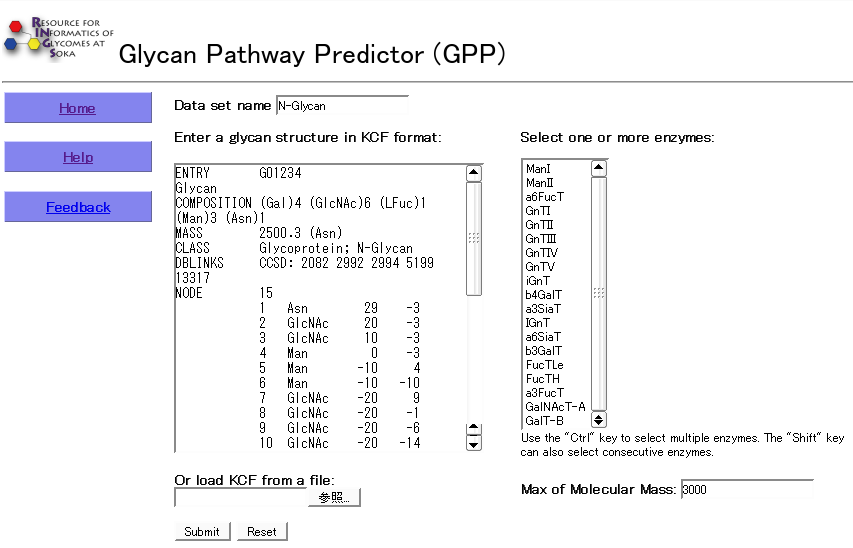
Usage
There is one major text field for entering a starting N-glycan structure in KCF format. This input can also be provided in a text file which can be loaded directly. A selection menu for the enzymes to work on the generated structures is given. Multiple enzymes may be selected simultaneously by using the shift- or alt- keys while clicking the enzyme name.
- Glycan structure information is required in KCF format as input data.
- Users can write sutructures directly or load a file.
- Users need to select at least one enzyme from enzyme list.
- Users need to specify the maximum molecular mass to produce in the pathway.
- A biosynthetic pathway is predicted by clicking the submit button.
-
Results
Each node represents a glycan structure in Linear Code format. A generated pathway is displayed with the ZGRViewer applet so that users can easily see the whole pathway as well as parts of the pathway at the same time.
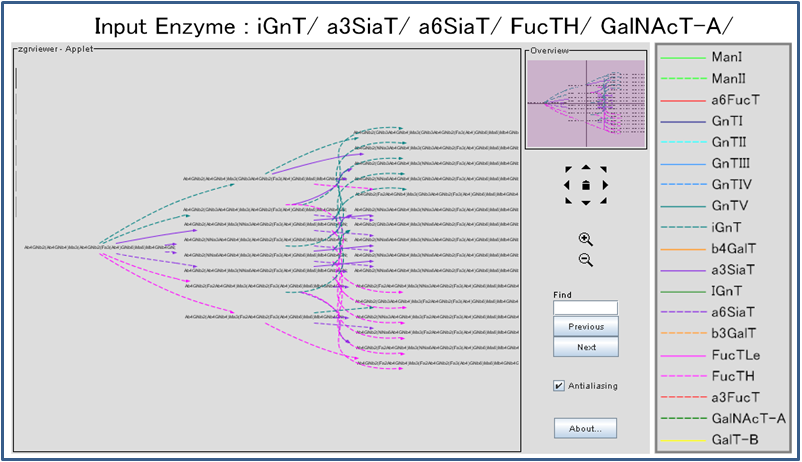
Double-clicking on a glycan structure will display the detailed information for the selected structure.
(i) Clicked glycan structure is used as an input glycan.
(ii) Predicted structure modified by enzyme (a) is shown in (b). Users can see the enzyme information by clicking an Interaction ID that starts with the letter "R".
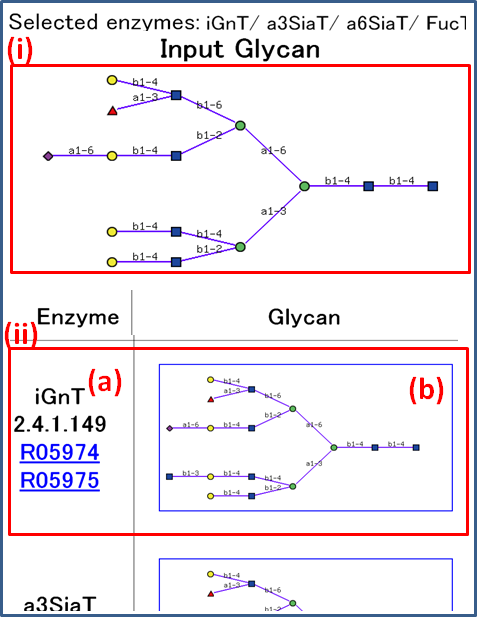
Clicking on the image of a generated structure (b), the tool will recalculate the possible enzymes from the original selection that may catalyze a bond on the clicked glycan, and a new results page will be displayed. Thus the biosynthesis pathway can be examined one structure at a time.
(Top)
ProfilePSTMM
-
Profile PSTMM Overview
Profile PSTMM is a probabilistic model (similar to hidden Markov models, HMMs), that can find patterns (profiles) in glycan structures that may not necessarily be easily found by the naked eye. It is implemented based on the work by Aoki-Kinoshita et al., 2006. In particular, it can predict longer-range patterns in the data, such as those between the non-reducing and reducing ends. Thus it is useful for predicting possible recognition patterns in glycans.
-
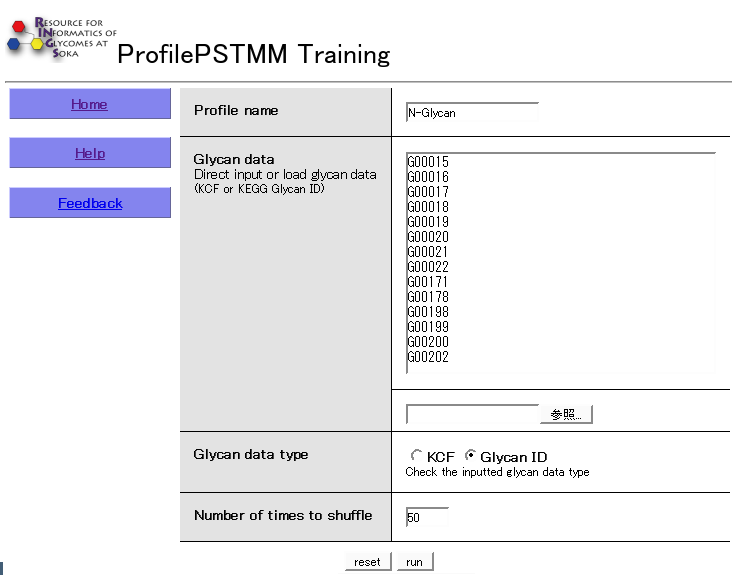
Usage
Since the profiles that are learned from the data are all stored for easy retrieval, the tool takes as input a Profile name that is unique. In the Glycan data section, glycan structure information can be specified using a list of KEGG Glycan IDs (beginning with the letter 'G' followed by five digits), one per line, or a list of structures in KCF format. If a list of IDs is given, an optional parameter for the 'strength' of the particular ID can be specified as a number following each ID. This may be useful when using Glycan Array data, for example, when the binding affinity of a particular glycan is stronger or weaker compared to others. The actual glycan data type specified should be selected, and the Number of times to shuffle option is used to indicate how many times the learning process should be repeated. This is useful because this model is a probabilistic model, whereby local optimums may be found. In order to ensure that the results contain a global optimum, the learning process should be repeated many times. Note, however, that increasing this number will also increase computation time.
-
Results
The results include a list of the learned profiles ranked according to their Likelihood scores, which is computed based on the learned parameters of the probabilistic model. The Glycan Profile is also displayed as a figure, where for each position, the probability of a particular monosaccharide is listed.
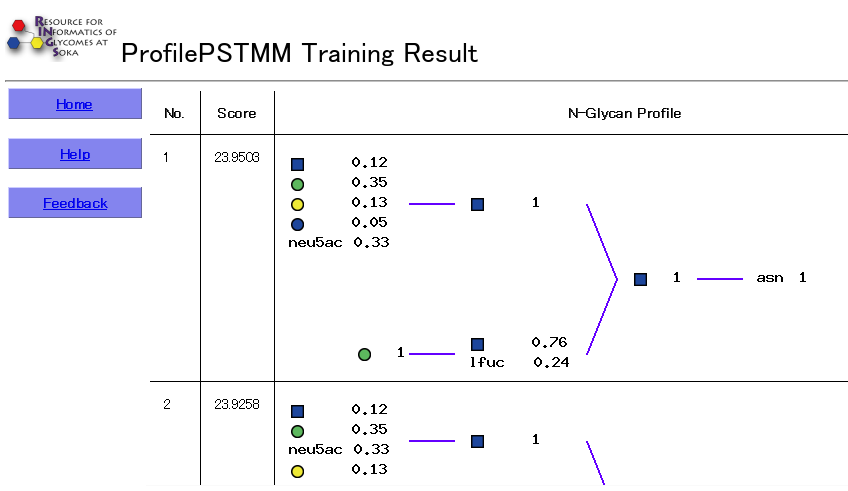
(Top)
Utilities
-
Utilities Overview
The RINGS Utilities consists of several useful tools for converting between various formats of glycan structures, including KCF, LINUCS, GlycoCT and IUPAC.
-
Usage
For most of the utilities, one or more glycan structures in the specified input format can be input either in the main text field or via the file browser. An option to display the results in HTML format, for ease of viewing, or in text format, for easy copy-and-pasting, is provided.
-
Results
The results are displayed in either of the selected output formats (HTML or text).
-
Example: KCF to LINUCS
First input one or more glycan structures in KCF notation and select whether to display the results in HTML or as text. Then click the "Get LINUCS" button.
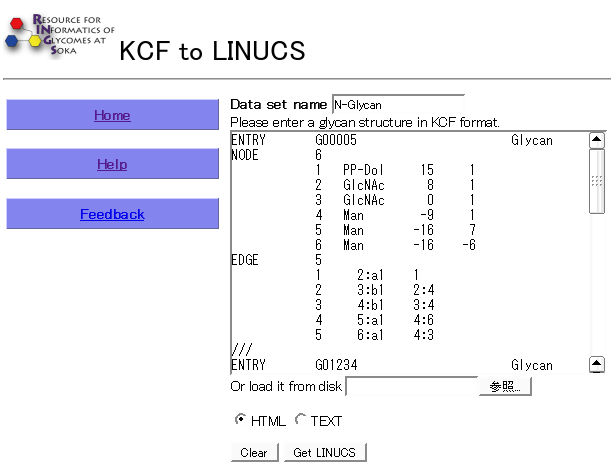
- Results
The following figure is a result of the KCF to LINUCS tool using default options.

The name of each input structure (as specified in the KCF file) is displayed next to the output LINUCS format.
(Top)
KCF to Mol
-
KCF to Mol tool overview
This tool is similar to the other utilities in that it converts a glycan structure between different formats.
However, it is different in that it produces an image as a result by which users can check the output before
downloading the output file.
-
Usage
- Users can write glycan strustures in KCF format directly or load a text file.
- Any of the options for JChemiPaint java applet, Jmol java applet and Chime plugin can be selected to display the results in the browser.
- Click View Mol button.
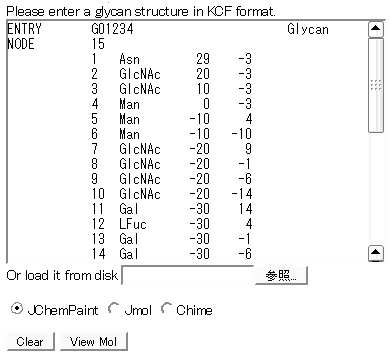
- Results
The results will display the resulting structure in the display form selected in the input, together with the glycan structure as a tree of monosaccharides. Examples of the different display forms are illustrated below.
JChemiPaint
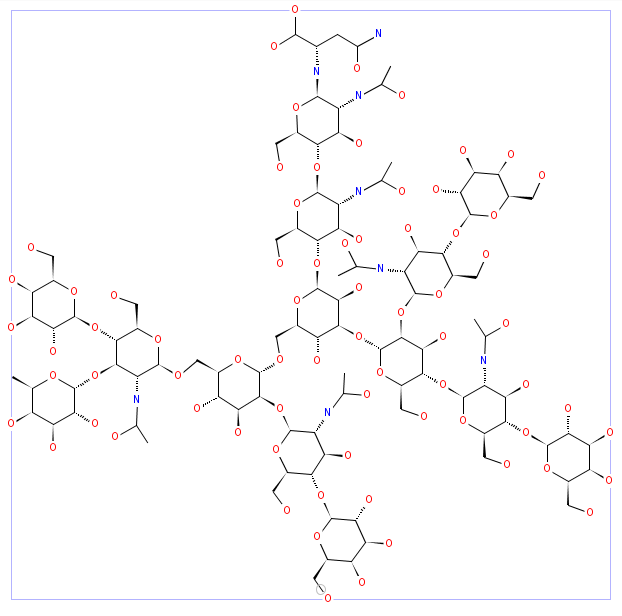
Jmol
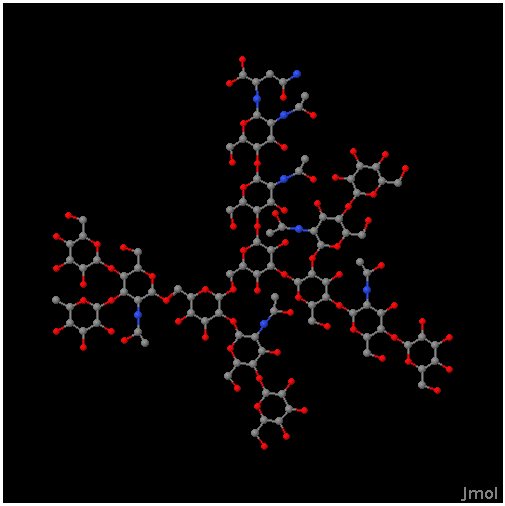
Chime
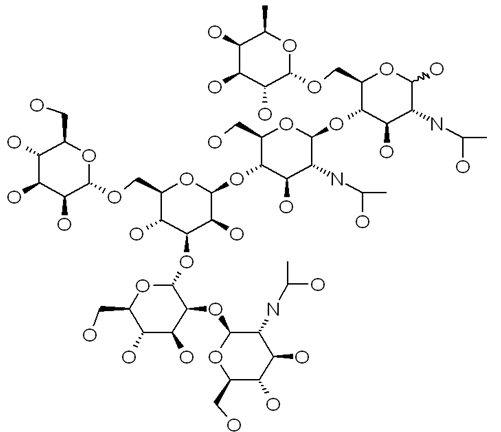
Clicking the "Get Molfile" at the bottom of the output page, the structure will be displayed in MOL format, which can be copy-and-pasted into any text file for local use.
Clicking the "KCFtoMol" link will return the user to the main page of this tool.
(Top)
User and data management
-
User and data management system overview
RINGS allows users to register such that data that has been analyzed can be stored on the server for
easy access and sharing. All of the corresponding results of the analysis tools in RINGS can also be
stored and retrieved later for further analysis.
-
Usage
- Click the "user registration" link at the top of the main page of RINGS.
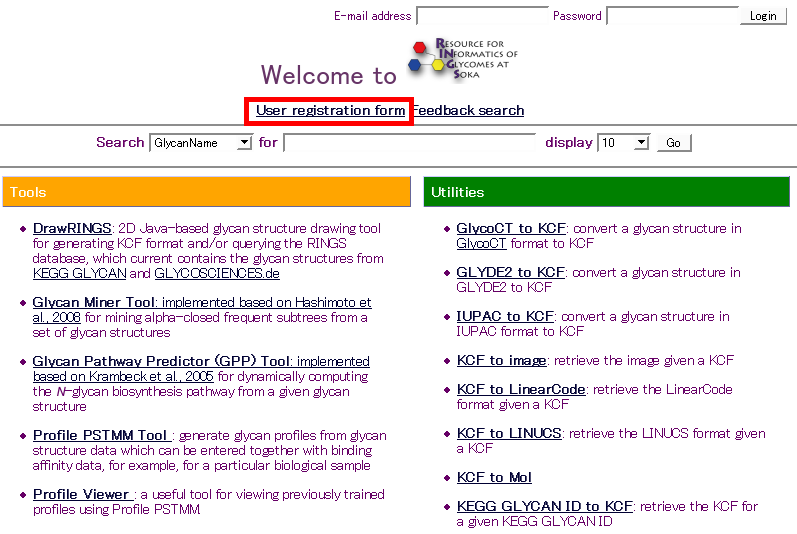
- Name, e-mail address, country and password are required for registration. When complete, click the "Submit" button.
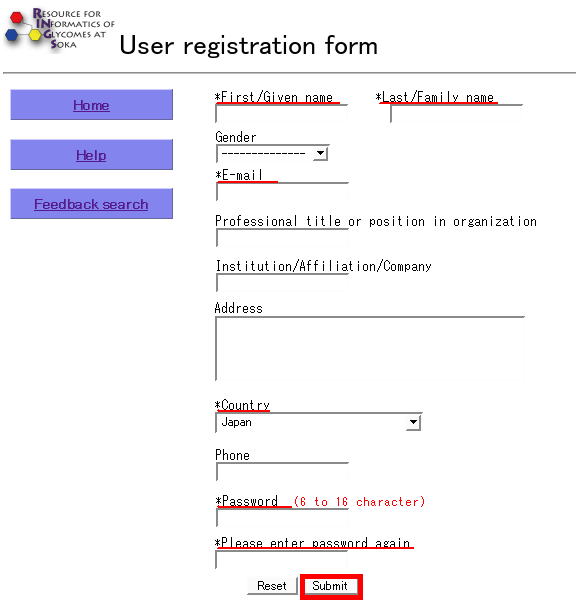
- Once registration is successful, the user can login by entering their e-mail address and password.

- To see how data is stored, select and execute a tool or utility.
- The analysis results will be automatically saved on the server, and the data set name will be displayed in the data tree.
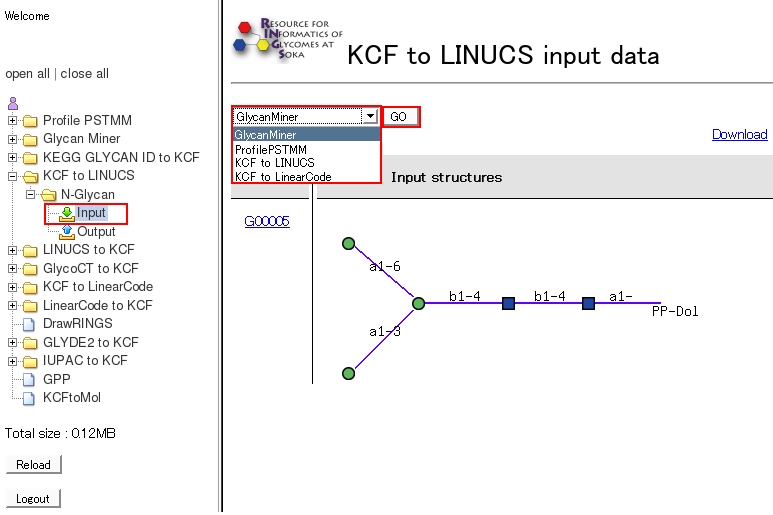
- By clicking on "Input" of a certain tool, the detailed information are displayed. Then, users can choose a tool from the pull-down menu, which will be displayed with the previous input data by clicking the "GO" button.
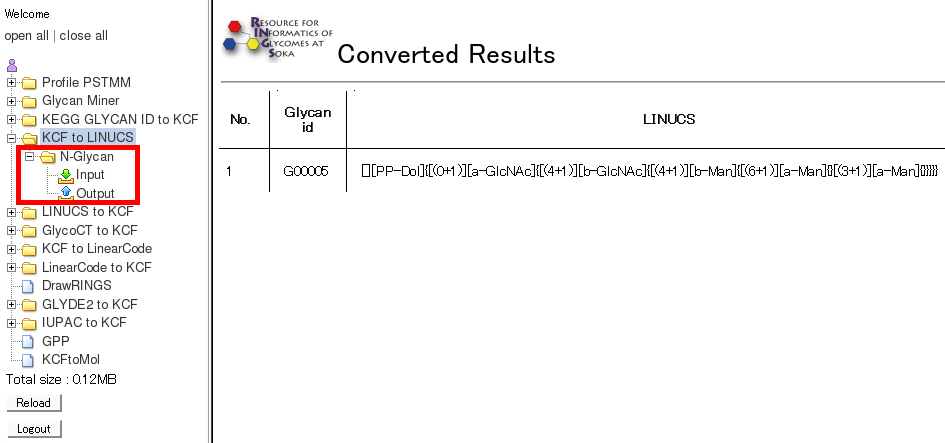
- By clicking on "Output" of a certain tool, the detailed infomation are displayed for all the past runs of the tool.
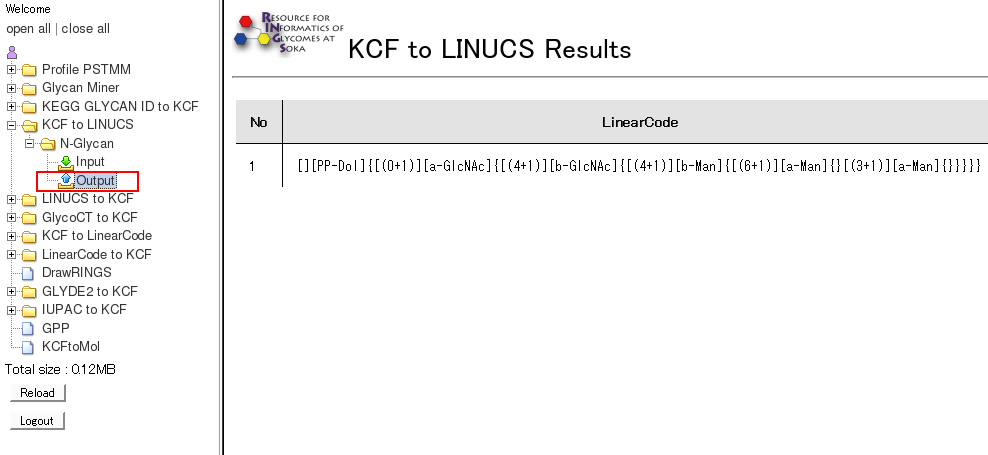
(Top)
Advanced search
-
About Advanced Search
Advanced search allows users to use multiple conditions for queries and more complex queries of the RINGS database.
For each condition, logic operators AND and OR can be used, and case-sensitive searches are also possible.
Details of the search parameters are provided below.
-
Advanced search page options
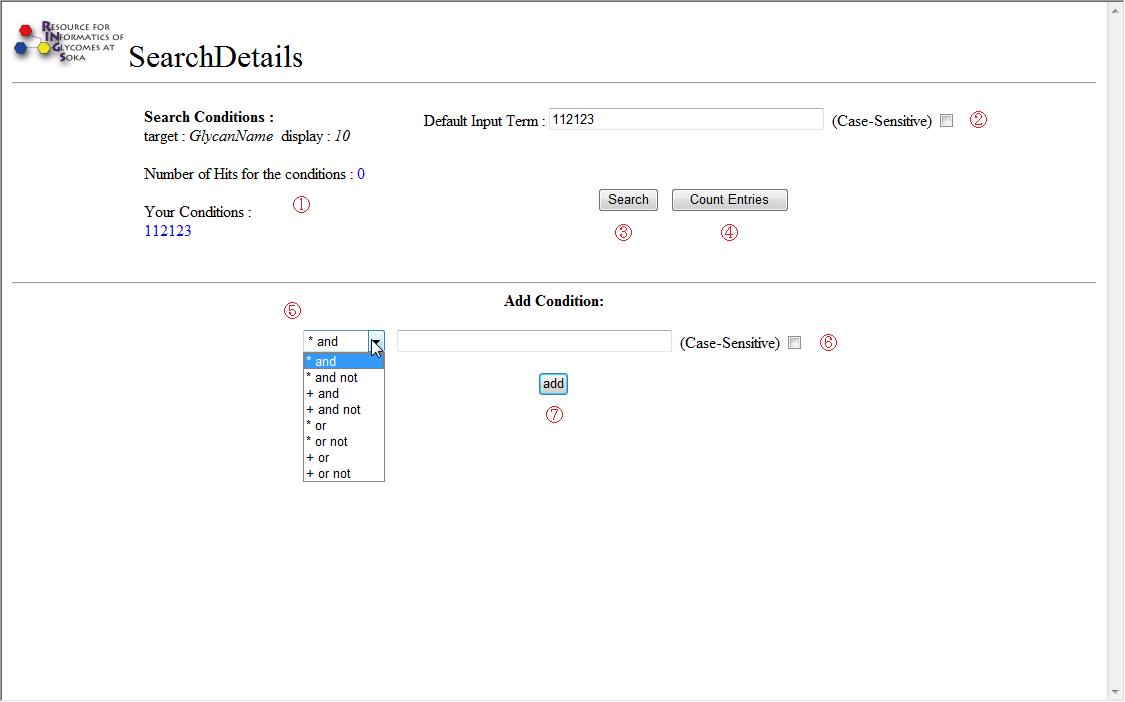
This figure is a snapshot of the default Advanced search page.
Each option numbered ① through ⑦ is described below.
-
Search conditions
Target:Selected database name
Display: Number of hits to display per page
Number of hits for the conditions:Number of hits found using the currently displayed options
Your conditions:A listing of the current search conditions
-
Default input term
This text field will contain the search terms that have already been inputted (if any). This can be modified as needed.
When the Case-Sensitive checkbox is checked, the search will be made distinguishing lower and upper case letters.
-
Search Button
When clicked, the current search options and parameters will be executed, and the resulting hits page will be displayed.
-
Count entries Button
This button can be used instead of the Search Button to retrieve only the number of hits that match the current conditions.
-
Logical operation (pull-down menu)
The operation by which to combine additional search conditions can be selected using this pull-down menu. The following eight options are available.
- * and
AND with the entire current search conditions.
- * and not
AND NOT with the entire current search conditions.
- + and
AND only with the immediately preceding search condition.
- + and not
AND NOT only with the immediately preceding search condition.
- * or
OR with the entire current search conditions.
- * or not
OR NOT with the entire current search conditions.
- + or
OR only with the immediately preceding search condition.
- + or not
OR NOT only with the immediately preceding search condition.
-
Additional search conditions
Similar to "Search conditions", additional search terms and case-sensitivity can be inputted here.
-
add Button
Additional search conditions can be added by pressing this button.
When this button is clicked, a page similar to the one below will be displayed.
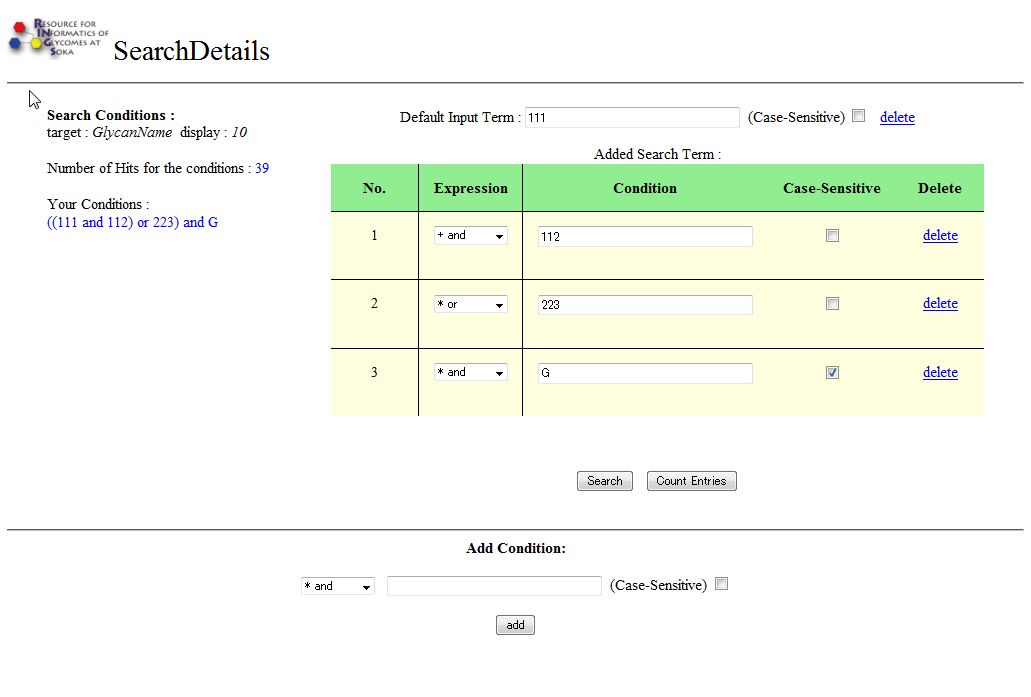
When the add button is pressed, the inputted search conditions will be added to the list above.
The number of Hits and a simplified listing of the existing search conditions ("Your Condition") will also be updated.
The "delete" link can be clicked on any existing condition to remove it from the search list.
-
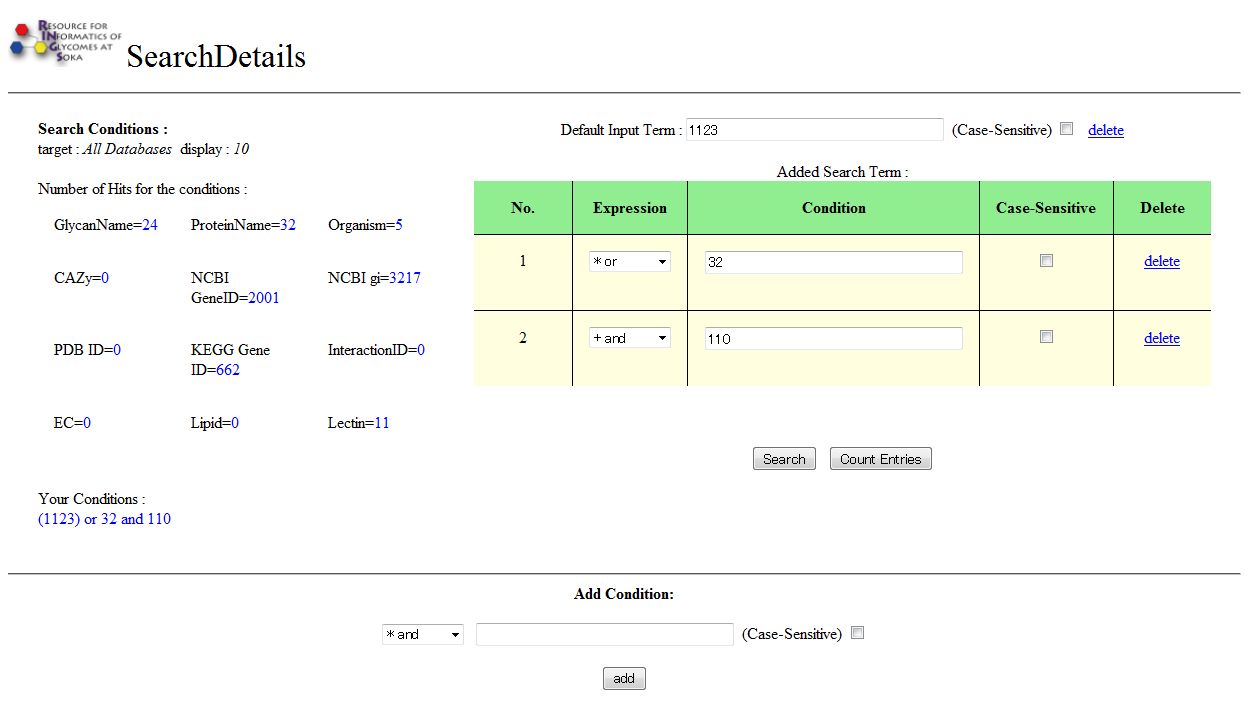
Advanced Search page for All Databases
When the Advanced Search is executed on the "All Databases" pull-down menu on the top page of RINGS, a slightly different page is displayed. For each database in RINGS, the number of Hits are displayed.
-
About the Results page
When the "search" button is pressed, the Results page is displayed, similar to the following.
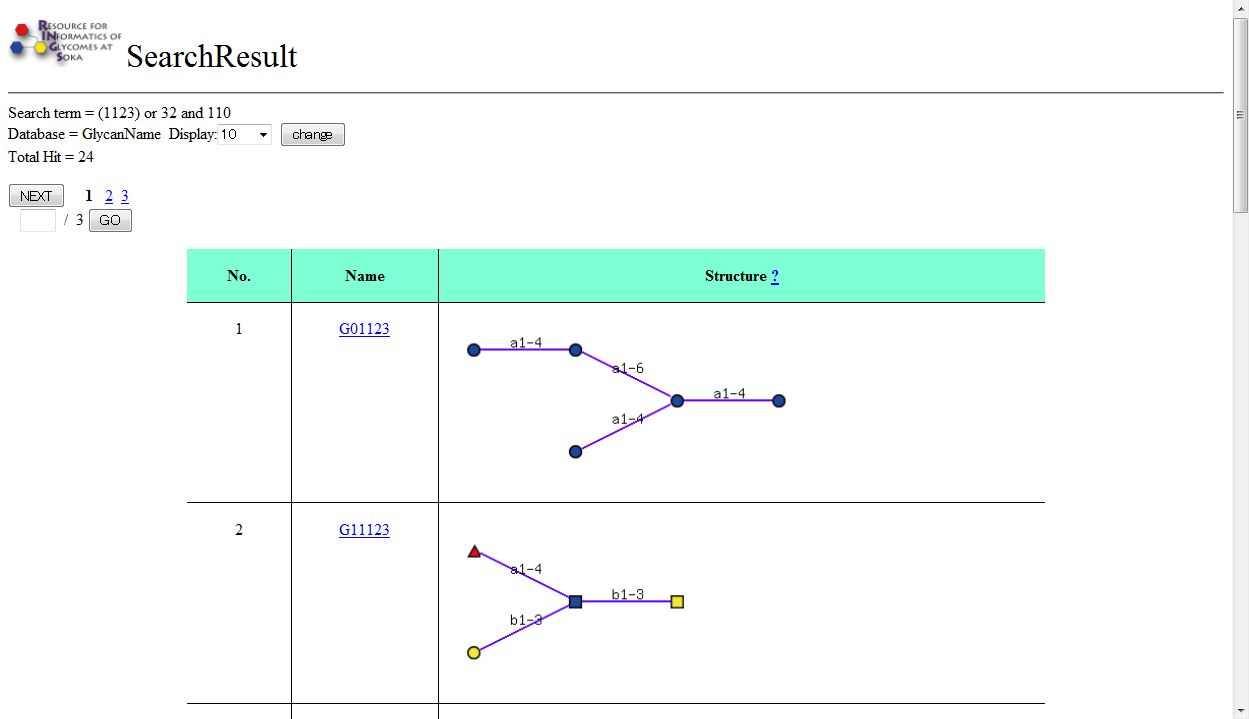
When "All Databases" is selected, the following page is first displayed, indicating the number of hits for each database. When a number is then clicked, the Results page will be displayed.

(Top)
|